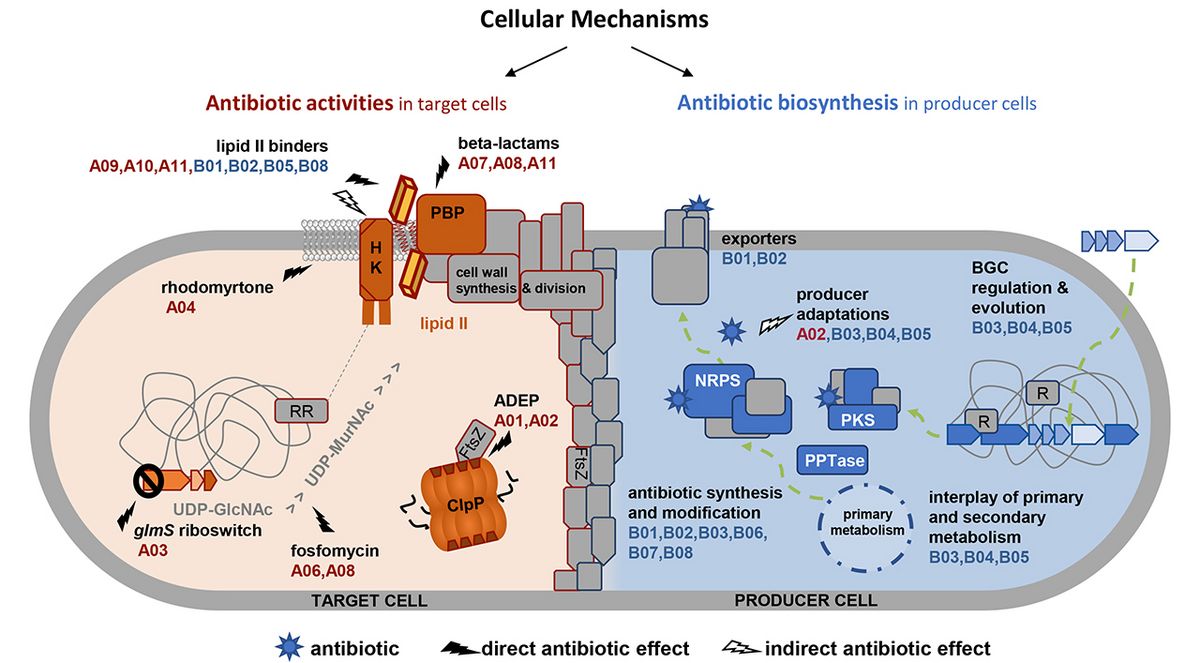
Project B01
Spatial and temporal cellular assembly of the glycopeptide biosynthetic complex
Balhimycin, a glycopeptide synthesized by Amycolatopsis balhimycina, is a heptapeptide, which is modified by cyclization, methylation, halogenation, and glycosylation. Using genetics and biochemistry, its biosynthetic steps have been elucidated indicating the occurrence of a large multi-enzyme complex that catalyzes amino acid assembly as well as various modifications. Whereas the chronology of the reactions could be defined, nothing is known about the positioning and dynamics of such a super-complex. Protein interaction studies and state-of-the-art microscopy will localize the complexes in the cell, identify further putative interaction partners, and define the temporal assembly of the complex.
Principal Investigators
Interfakultäres Institut für Mikrobiologie und Infektionsmedizin
Mikrobielle Wirkstoffe / Biotechnologie
Eberhard Karls Universität Tübingen
Auf der Morgenstelle 28, 72076 Tübingen
+49 (0)7071 29-76944
wolfgang.wohllebenbiotech.uni-tuebingen.de
Interfakultäres Institut für Mikrobiologie und Infektionsmedizin
Mikrobielle Wirkstoffe / Biotechnologie
Eberhard Karls Universität Tübingen
Auf der Morgenstelle 28, 72076 Tübingen
+49 (0)7071 29-78840
evi.stegmannbiotech.uni-tuebingen.de
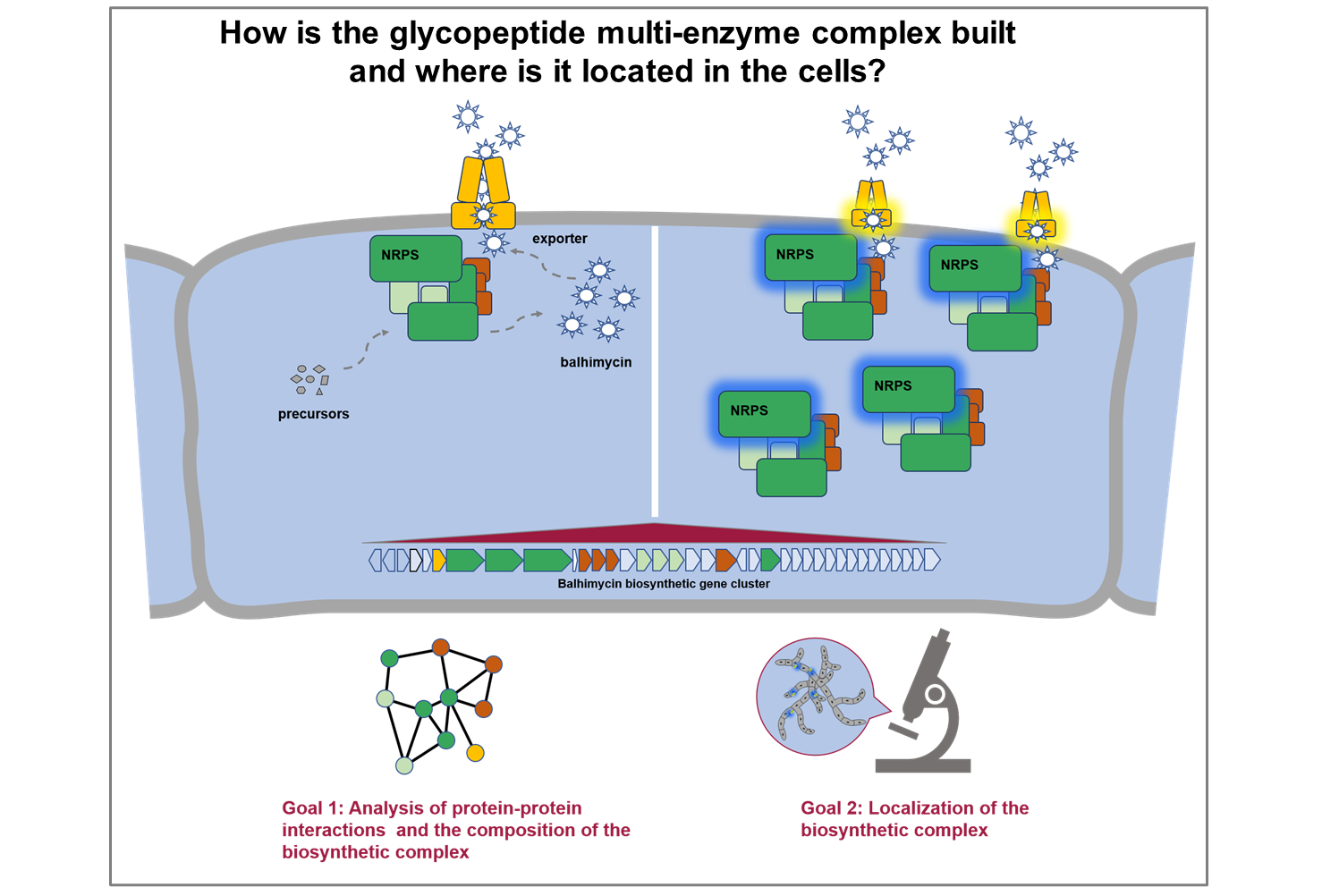
Project B02
Functional dissection of transport processes during glycopeptide antibiotic production
Bacterial biosynthetic gene clusters of natural compounds – like antibiotics – mostly encode exporters of the compounds produced. Amycolatopsis balhimycina, producer of the glycopeptide antibiotic balhimycin, encodes the ABC exporter Tba, which was shown to be critical for balhimycin secretion but not for self-resistance of the producer strain. Here, we want to further characterize the role of Tba in balhimycin production and assess its specificity towards glycopeptide antibiotics. We will employ an integrated approach using bacterial genetics, targeted analysis of secondary metabolites, protein localization, and protein-protein interaction analysis as well as in vitro transport assays.
Principal Investigator
Interfakultäres Institut für Mikrobiologie und Infektionsmedizin
Zelluläre und Molekulare Mikrobiologie
Eberhard Karls Universität Tübingen
Elfriede-Aulhorn-Str. 6, 72076 Tübingen
+49 (0)7071 29-84238
samuel.wagnermed.uni-tuebingen.de
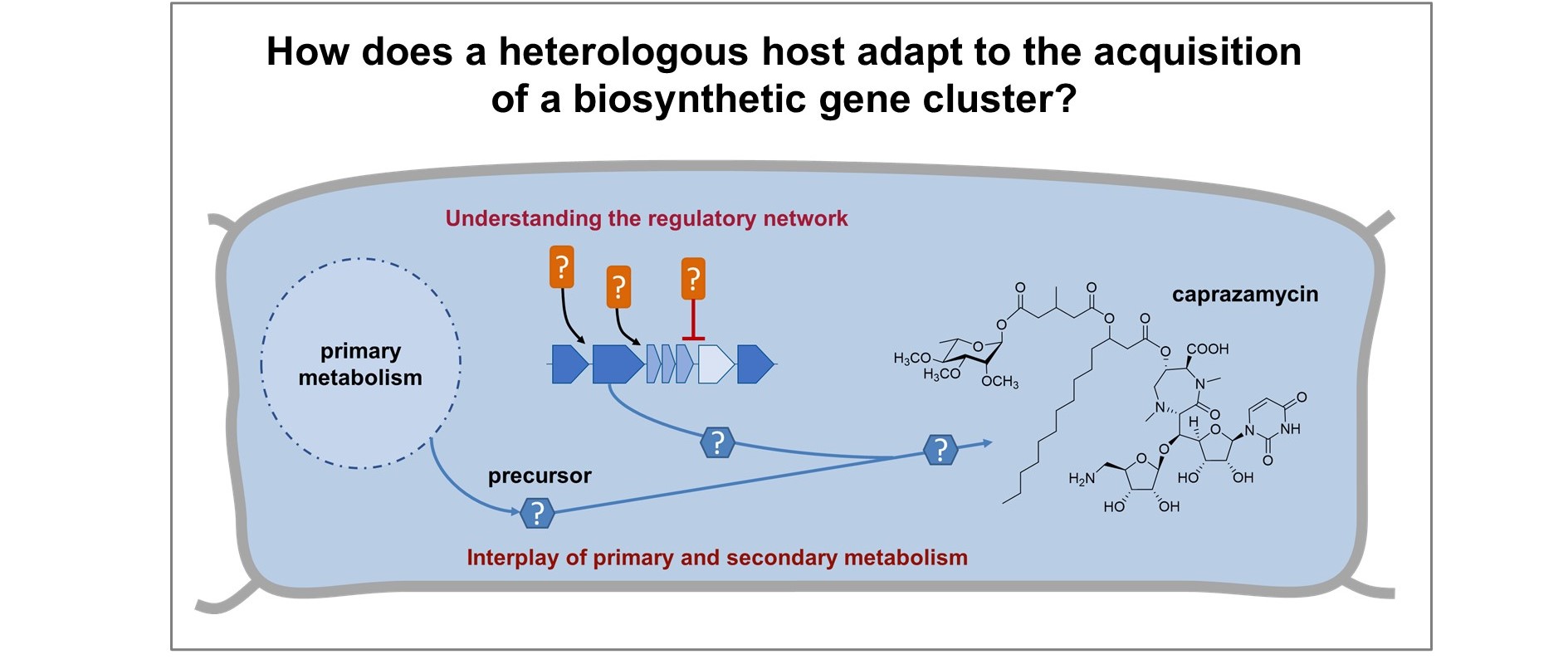
Project B03
Cellular responses to heterologously expressed antibiotic gene clusters
This project will address the cellular responses and adaptations of the common host strain Streptomyces coelicolor to heterologously expressed gene clusters. Surprisingly little is known about what actually happens in a heterologous strain during natural product biosynthesis, besides the occurrence or non-occurrence of the desired compound. It will be studied if the novobiocin and caprazamycin gene clusters respond to global regulators of the heterologous host, how biosynthetic routes offered by the host affect precursor supply, and how these precursors are finally incorporated into the desired compound.
Principal Investigators
Pharmazeutisches Institut
Pharmazeutische Biologie
Eberhard Karls Universität Tübingen
Auf der Morgenstelle 8, 72076 Tübingen
+49 (0)7071 29-75483
bertolt.gustuni-tuebingen.de
Prof. Dr. Leonard Kaysser
Institute for Drug Discovery
Department Pharmaceutical Biology
University of Leipzig
Eilenburger Str. 14, 04317 Leipzig
+49 341 97-11851
leonard.kaysserpharm.uni-leipzig.de
Project B04
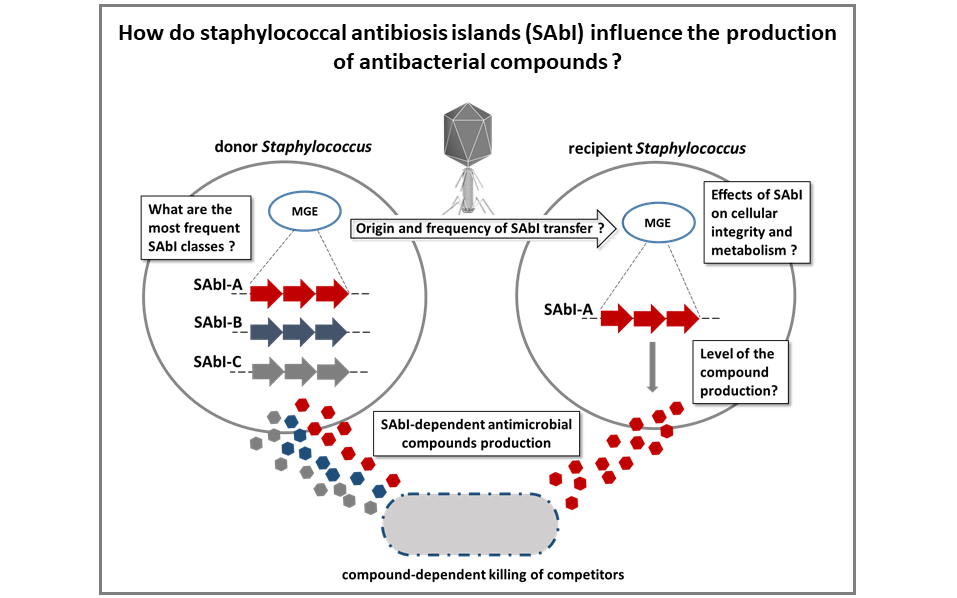
Horizontal transfer, evolution, and cellular integration of Staphylococcus antibiosis islands (SAbIs)
We recently described that staphylococcal species produce a variety of antibacterial compounds. The responsible gene clusters show signs of mobility and we suggest to term them SAbIs. We will identify families of SAbIs and investigate the routes and frequency of SAbI transfer. Using in vitro evolution experiments and metabolomics/genomics approaches, we will investigate adaptation processes that mediate producer resistance and metabolic fluxes that optimize fitness in SAbI recipients. The project will deliver novel insights into physiologic and metabolic bottlenecks as well as associated adaptations, shaping the diversity of compounds produced within bacterial communities.
Principal Investigators
Interfakultäres Institut für Mikrobiologie und Infektionsmedizin
Infektionsbiologie
Eberhard Karls Universität Tübingen
Auf der Morgenstelle 28, 72076 Tübingen
+49 (0)7071 29-75938
simon.heilbronneruni-tuebingen.de
Interfakultäres Institut für Mikrobiologie und Infektionsmedizin
Infektionsbiologie
Eberhard Karls Universität Tübingen
Auf der Morgenstelle 28, 72076 Tübingen
+49 (0) 7071 29-75935
andreas.pescheluni-tuebingen.de
Project B05
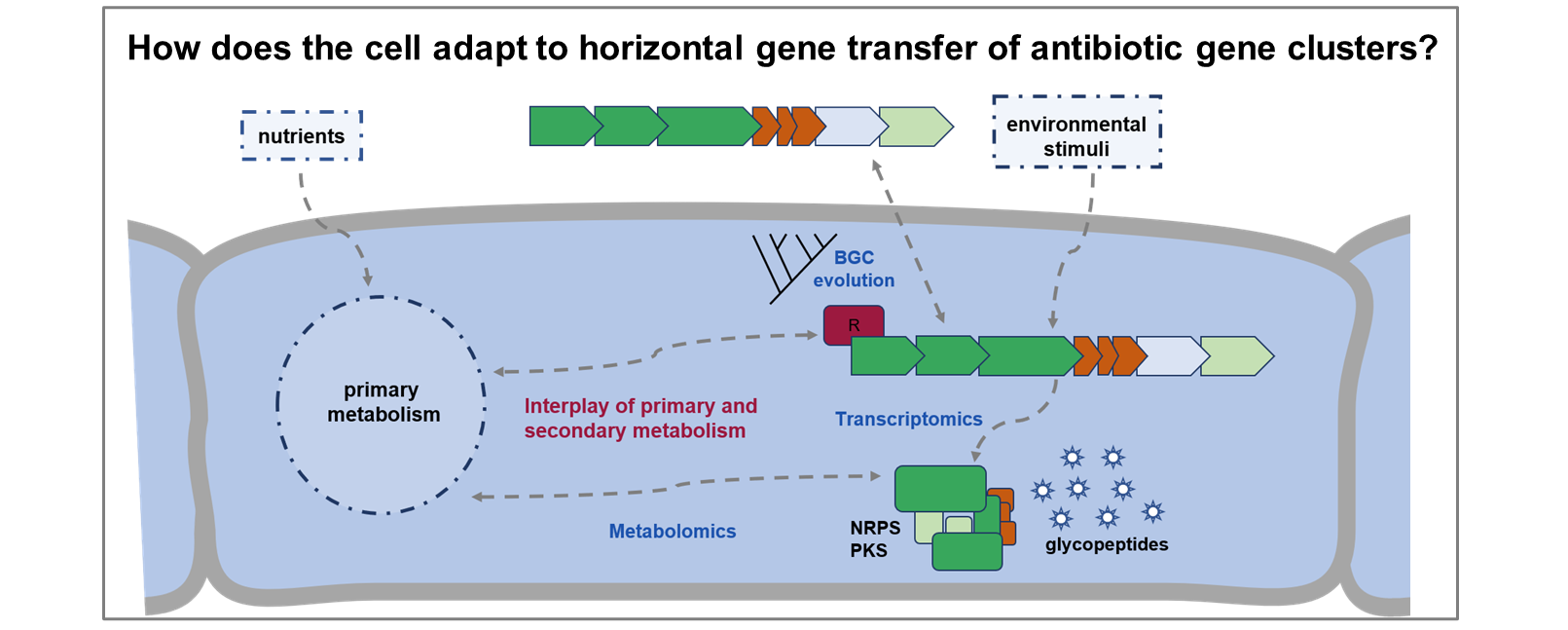
Cellular adaptation mechanisms to horizontal gene transfer of antibiotic gene clusters in the model system Amycolatopsis
Up to 80% of secondary metabolite gene clusters were relatively recently acquired via horizontal gene transfer. However, not much is known about the specific adaptations a cell has to go through in order to keep the cellular processes functioning and synchronized. We want to understand the effects and adaptation mechanisms of the bacterial cells that acquire antibiotic gene clusters and identify key regulatory mechanisms to control and fine-tune the equilibrium of primary and secondary metabolism. As a model system, we chose glycopeptide antibiotics in the genus Amycolatopsis.
Principal Investigators
Interfakultäres Institut für Mikrobiologie und Infektionsmedizin
Applied Natural Products Genome Mining
Eberhard Karls Universität Tübingen
Auf der Morgenstelle 28, 72076 Tübingen
+49 (0)7071 29-78841
nadine.ziemertuni-tuebingen.de
Interfakultäres Institut für Mikrobiologie und Infektionsmedizin
Mikrobiologie / Biotechnologie
Eberhard Karls Universität Tübingen
Auf der Morgenstelle 28, 72076 Tübingen
+49 (0)7071 29-78840
evi.stegmannbiotech.uni-tuebingen.de
Project B06
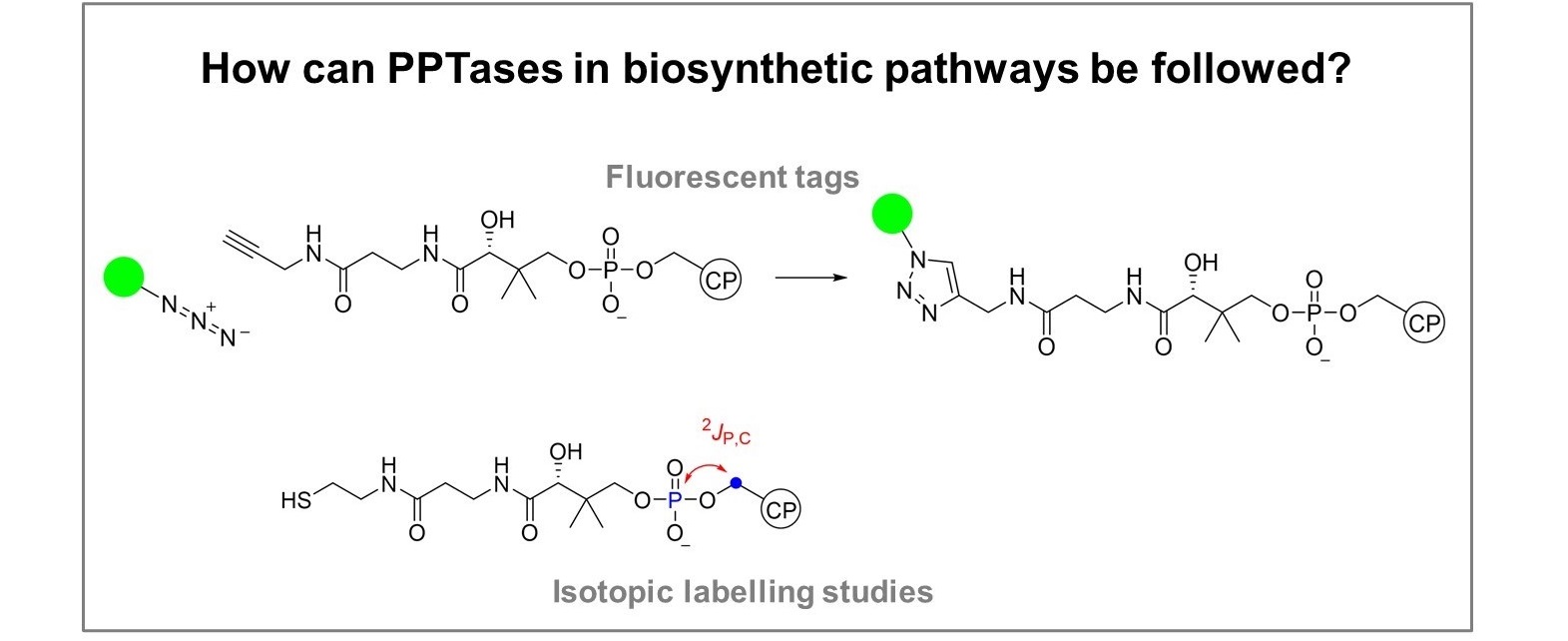
Phosphopantetheinyl transferases in antibiotic biosynthesis
Phosphopantetheinyl transferases (PPTases) will be studied in the polyketide-synthase/non-ribosomal peptide synthetase (PKS-NRPS) for antimycins and blastmycinones in Streptomyces ambofaciens and in the trans-AT-PKS for bacillaene in Bacillus amyloliquefaciens. Antimycin and blastmycinone production depends on two PPTases, suggesting that blastmycinone biosynthesis may be independent of antimycin biosynthesis by only partial activation of the PKS-NRPS by one PPTase. This hypothesis will be challenged experimentally. New sensitive methods will be developed that allow to follow the PPTase reaction. Isotopic labeling experiments will be performed for structure elucidation.
Principal Investigator
Kekulé-Institut für Organische Chemie und Biochemie
Rheinische Friedrich-Wilhelms-Universität Bonn
Gerhard-Domagk-Straße 1, 53121 Bonn
+49 (0)228 73-5797
dickschatuni-bonn.de
Project B07
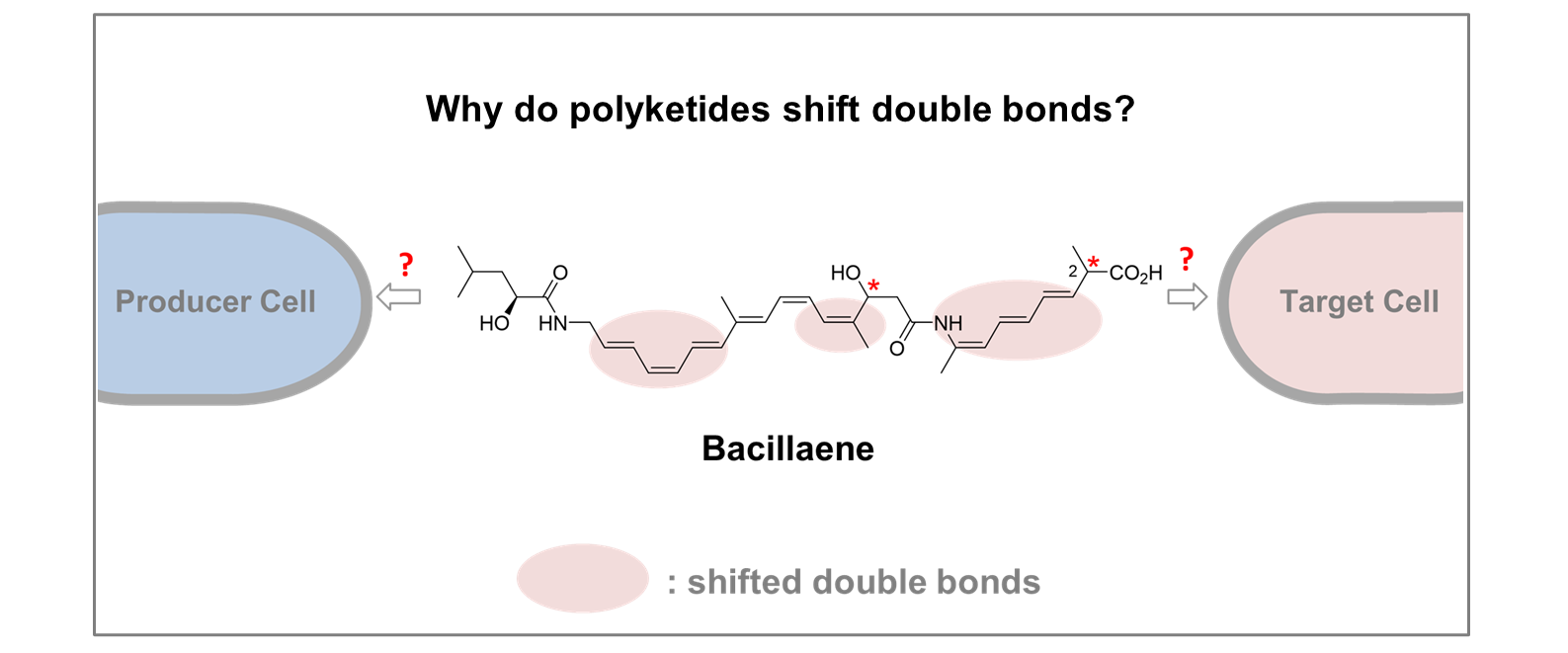
Effects of double bond shifts in polyketide biosynthesis
A case study with bacillaene, a highly potent but extremely labile polyene polyketide of bacterial origin, will be pursued. Different double bond shift and isomerization mechanisms during and after biosynthesis and general controlling elements for these processes will be studied in detail to understand their effects on the producer as well as on the target cell. The natural product, selected biosynthetic intermediates thereof as well as designed analogs will either be isolated and analyzed or prepared by first total syntheses.
Principal Investigator
Kekulé-Institut für Organische Chemie und Biochemie
Rheinische Friedrich-Wilhelms-Universität Bonn
Gerhard-Domagk-Str. 1, 53121 Bonn
+49 (0)228 73-2653
dirk.mencheuni-bonn.de
Project B08
Structures of cellular machineries relevant for antibiotic synthesis and action
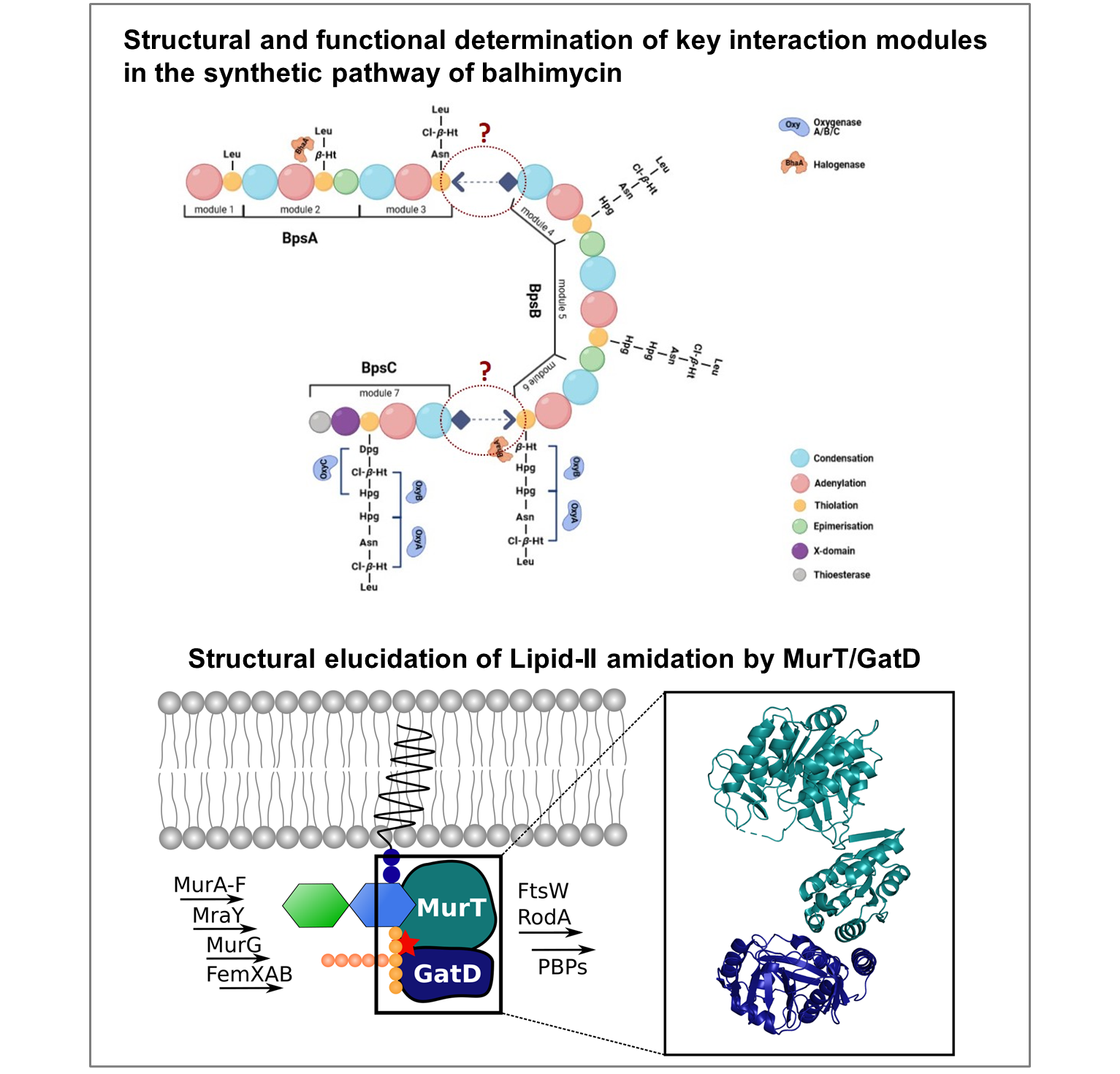
In the first part of this project, we will characterize the interactions between subunits of a multi-enzyme complex that synthesizes the glycopeptide antibiotic balhimycin in Amycolatopsis balhimycina. In the second part, we will identify substrate specificities and enzymatic mechanisms of enzyme complexes that catalyze lipid II amidation in Staphylococcus aureus and A. balhimycina. Both objectives will be pursued using protein crystallography and associated biophysical techniques as well as functional assays. The overall goal of the proposed work is to advance an understanding of enzymatic mechanisms relevant for the biosynthesis and function of antibiotics.
Principal Investigator
Interfakultäres Institut für Biochemie
Applied Natural Products Genome Mining
Eberhard Karls Universität Tübingen
Hoppe-Seyler-Str. 4, 72076 Tübingen
+49 (0)7071 29-73043
thilo.stehleuni-tuebingen.de
- Project B01
- Project B02
- Project B03
- Project B04
- Project B05
- Project B06
- Project B07
- Project B08